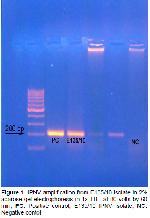
Infectious pancreatic necrosis virus is highly
prevalent in rainbow trout cultured in cage culture
systems in the Eastern Anatolia region
13. One of the
main problems in epidemiological studies of IPNV is the
difficulty of typing new isolates due to the large range of
serotypes exist. A variety of IPNV isolates from around
the world are currently known, and there is great interest
in classifying them. Various techniques have been used
to achieve this goal, the first being serology
14 and
restriction fragment length polymorphisms
15,16.
Presently, the most commonly used technique to
determine single-nucleotide changes between
sequences has been the sequencing of several IPNV
fragments of genome segments A and B
17,18. In this
study, it was determined nucleotide sequence VP2
region of IPNV isolate (Figure
1). The result of this study
has shown that overall, the KC525214 ELAZIĞ isolate
was identical to and clustered with the Norwegian,
French, Danish, Taiwanese, Spanish and American
isolates into one genogroup closely related to the VP2
reference sequence (95.1-96.6%). Norwegian IPNV
isolates were generally the most similar to KC525214
ELAZIĞ (96.6%) (Table
2). In contrast, all the England,
Canada and Japan reference sequences were more
distant. Isolate used in the present study belong to one
genogroup equivalent to genogroup 5 proposed by Blake
et al.
10 and are therefore consistent with the A2
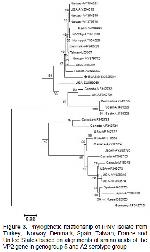
serotype (Figure
2A,B; Figure
3).

Büyütmek İçin Tıklayın |
Table 2: Comparison of infectious pancreatic necrosis virus (IPNV) KC525214 ELAZIĞ isolate, showing percentage
nucleotid sequence identity matrix based on sequences of the VP2* gene in previously reported isolates in A2 serotype |
Currently, the most common system for analyzing the
genetic diversity of a virus and classifying virus
genogroups is through genomic sequencing of genes
that generally code for the virion surface proteins or that
encode the virus polymerase. For IPNV, several authors
have used the sequences of nucleotides and amino
acids to make such classifications based on the VP2
region or the entire ORF of segment A15,16,19,20
some of authors are also used for genogrouping IPNV
based on VP118,21 or the VP3 and VP5 genes21.
In this study, we used the sequence of nucleotides 20-
225 of genome segment A, which constitute a fragment
of VP2 (Figure 3). The genogroups in the phylogenetic
tree are in concordance with the classification proposed
by Blake et al.10.
Infectious pancreatic necrosis virus was first reported
from rainbow trout by Candan22 in Turkey and after
that was investigated by Ogut and Altuntas24,
Albayrak and Ozan23, Gürçay et al.13 IPNV is now
endemic and factors supporting endemicity are not fully
explored at present in Turkey. It was reported that the
severity of IPNV in rainbow trout depends on species,
strain and age of the fish25,26 along with the two key
disposing factors, stress and temperature24.
The result of sequence analysis in this study implied
that the origin of IPNV isolated from trout farms in the
Eastern Anatolia was the hatcheries where
Oncorhynchus mykiss eggs transported from Norwegian,
French, Danish, Taiwanese, Spanish and American
isolates (Figure 3). The findings strongly indicate that
trout become infected in the hatcheries in the
broodstock, and then the infection becomes latent. This
result also suggests that probably IPNV, highly
contagious leading to rapid spread in the region, became
endemic in the region due to the transfer of latently
infected fish and reproductive fluids. American serotype
that was transferred to Spain by the importation of
rainbow trout eggs from North American farms17.
Similarly, as occurred with the importation of IPNV Eastern Anatolia region, study has shown that the
KC525214 ELAZIĞ isolate is closely related to the
Norwegian, French, Danish, Taiwanese, Spanish and
American isolates.
KC525214 ELAZIĞ isolate used in the present study
belong to one genogroup equivalent to genogroup 5
proposed by Blake et al.10 and are therefore
consistent with the A2 serotype. In the other study,
nucleotide sequences of the VP2/NS region of IPNV
showed that all isolates collected in the Black Sea region
and surrounding areas were determined belonged to the
genogroup III Ogut and Altıntas27.
In conclusion, the findings presented here support
the view that the transfer of breeding materials is
associated with the transmission of pathogens. To
prevent the introduction of the infectious diseases into a
recirculating system, the best recommendation is to
hatch eggs at the facility or buy fingerlings from a
certified disease-free source. Additionally, newly arrived
fingerlings should be quarantined before introduction into
the system and reared in a pathogen-protected
environment.
Acknowledgments
The authors would like to thank Tarımsal Araştırmalar
ve Politikalar Genel Müdürlüğü (TAGEM) for its financial
support.